Accelerated Data Processing Strategies
Vincent Brandon, Data Coordinator
January 13, 2021

Data preparation is an often overlooked part of data science and analytics. In-database has been the moniker for fast operations in large datasets. More recently, frameworks in python like Dask and Numba have made handling large datasets much easier and in some cases much more performant. Simple operations like summing a column of data across millions or billions of records can be made orders of magnitude faster by breaking the task into pieces and running it in parallel. Everything from basic statistics to joins traditionally done on CPUs in relational databases can be offloaded to in memory operations on specialized hardware. RAPIDS, built by Nvidia, exposes drop-in replacements that magically whisk gigabytes of data off to powerful GPUs for mind-boggling performance increases.
Why does the UDRC want accelerated data processing? Playing with cool tech is certainly up there, but there are cost savings to consider - not to mention the sanity of our researchers and analysts. The UDRC maps data to individuals across datasets from different government organizations. Determining the quality of data, the probability of a match, and then writing all that data out, is fine if the tables are a few thousand or even a hundred thousand rows each. But imagine checking a million rows against ten million possible people:
1,000,000 X 10,000,000 = 10,000,000,000,000 (A really big number).
Some operations are simply intractable without blocking out subsets of data to avoid this dreaded worst-case cross join every time we bring in new data. Before we calculate the probability of a match in a ten million row table, we need to search the space of individuals, for likely candidates, up to a million times (not really, there are tricks to lower that significantly). Seems crazy, but if we can reduce the number of possible matches to say 10 candidates for each row, then we only have ten million inferences to run. Better than ten trillion by a mile. In a GPU, this takes seconds. In a shared database, simply communicating this many indexing functions is a chore. We don’t want to write custom database code for every table and deal with poor performance. Enter RAPIDS.
For those who’ve worked with the Pandas library in python, RAPIDS’ cudf will feel very familiar. We can generate some fake data, just letters and numbers, and see how the performance scales without custom optimization (both could probably be faster).
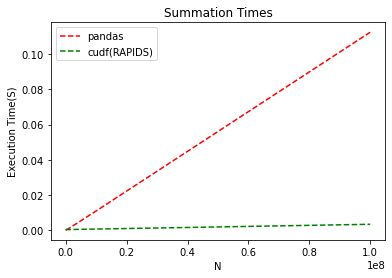
We can see that summing rows in Pandas is fast. 10 million rows completed in less than a tenth of a second on the CPU. Looking at RAPIDS, the task takes…a tenth of that, and never reaches a hundredth of a second. What’s amazing here is that not only is RAPIDS fast for smaller datasets, about as fast as Pandas, but as the dataset size increases, the slope stays near flat on the GPU.
At the UDRC, we do a lot of grouping to determine distinct names, count by gender, and more. Group by and count are staples of SQL. Pandas offers functionality that will perform similarly to a database in memory. What if we accelerate the process with RAPIDS?
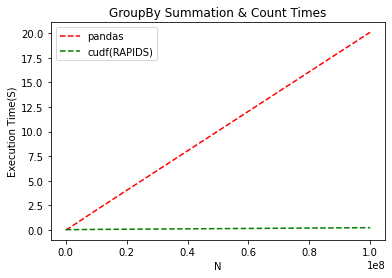
The improvement is staggering. The graph looks similar to summation, but pay attention to the scale on the Y-axis. On CPU, an operation that takes 20s doesn’t even take 1 on GPU. With RAPIDS, we see a 20X improvement on an operation we need to do dozens of times during record linkage. This is the difference between starting the program and coming in the next day to see results versus clicking ‘go’ and having it complete over a coffee break.
Getting started with RAPIDS is easy. Follow instructions on https://rapids.ai/start.html. At the time of this writing, Google Colab and BlazingSQL are by and large the best way to get started with RAPIDS. Workstation and production setups are still tricky. CUDA is still unstable in WSL 2 environments for windows users. Docker containers may have incompatibilities with host Nvidia drivers in some Linux environments.
View the notebook with examples on Github.